Journal of Multidisciplinary Dental Research
Volume: 4, Issue: 2, Pages: 25-30
Original Article
Sanju Somaiah1, Sumitha Ann David2, Balakrishna Shetty3, Roopa Siddegowda4
1Professor, Coorg Institute of Dental Sciences, Virajpet
2Sr Lecturer, Educare Dental college, Kottakkal
3Professor, Coorg Institute of Dental Sciences, Virajpet
4Sr Lecturer, Coorg Institute of Dental Sciences, Virajpet
Corresponding
Sanju Somaiah
Coorg Institute of Dental Science, K.K. Campus, Maggula P.O, Virajpet, Coorg
Phone : 9945527999
E-mail: [email protected]
Received Date:05 September 2018, Accepted Date:22 September 2018, Published Date:30 September 2018
Aims:
Settings and Design:
Methods and Material: Sample consisted of twenty subjects. 2 ml blood sample was taken using a 5 ml syringe and stored with anticoagulant, in an ice box. It was sent to the Laboratory within 24 hrs, for the SNP study. Restriction Fragment Length Polymorphism (RFLP) was done to check for variation in the MTHFR and MSX1 genes and the results were computed after statistical analysis.
Statistical analysis used: Descriptive statistics, Crosstabs (Contingency coefficient test), Chi-Square Test
Results:
Conclusions:
It was observed that polymorphism (gene mutation) was seen in MTHFR gene and not seen in MSX1 gene, for the present sample.
It was concluded that, although the polymorphism in MTHFR gene was not significant enough, MTHFR gene played a more prominent role than MSX1 gene. So this showed that MTHFR gene could be considered as a better marker in the development of NSCLP
Key-words: Genetics, MTHFR, MSX1, Cleft Lip/Palate
Key Messages: MTHFR gene plays a more prominent role than MSX1 gene in the development of Nonsyndromic Cleft Lip/Palate. Knowledge in genetic implications along with future advances in gene therapy can help prevent Clefts.
Cleft lip and palate, or isolated cleft lip or palate (CL/P) is a common congenital anomaly. The approximate incidence of cleft lip and palate is 1.4 per 1,000 live births in India. On an average, Cleft of the lip, palate, or both has a birth prevalence rate ranging from 1/1000 to 2.69/1000 amongst different parts of the world.
Clefts arise during the fourth stage in embryonic craniofacial development. The exact location of the cleft is determined by the location at which fusion of various facial processes failed to occur, due to some interference in the development.1
With a birth prevalence of approximately two per 1,000 livebirths, orofacial clefts rank among the most common birth defects in humans.2
Both genetic and environment factors are known to play a role in the development of oral clefts. Approximately 70 % of cases are nonsyndromic and 30% are syndromic. Some of the teratogens that affect the dentofacial development are Aspirin, Cigarette smoking, Dilantin, 6-Mercaptopurine and Valium. Several candidate genes have been proposed as risk factors by intense research and advances in genotyping technology. Some of these are MTHFR, MSX1, TGF-A, TGF-B, PAX9, RARA and DLX2.3
MTHFR gene provides instructions for making an enzyme called 'methylenetetra hydrofolate reductase', which is important in folate metabolism.4 This enzyme plays a role in processing of amino acids, the building blocks of proteins. It is important for a chemical reaction involving forms of Vitamin-B. The polymorphism in the MTHFR gene has been suggested as a risk factor for CL/P.
MSX1 gene is officially named as 'MSH homeobox 1'. It provides instructions for making a protein that regulates the activity of other genes. It acts during early development, to control the formation of many body parts. It is critical for the normal development of teeth and other structures in the mouth. It plays an important role in epithelial-mesenchymal interactions during craniofacial development. Significant linkage disequilibrium has been found between CL/P and MSX1 gene.
Both genes are associated with the processing of proteins, the building blocks of our body. Several studies have shown a possible association of MTHFR and MSX1 genes in the etiology of nonsyndromic CL/P.
The objective of our study was to find further evidence associating MTHFR and MSX1 gene in the etiology of NSCLP.
Subjects and Methods:
The sample consisted of 20 subjects (upto 17 years) reporting to the cleft clinic of Department of Orthodontics and Dentofacial Orthopaedics, Coorg Institute of Dental Sciences, Coorg, India. Informed consent was obtained from patients/parents of all subjects. The study was carried out after due approval from institutional ethical committee, and guidelines from Helsinki declaration were followed. A thorough clinical examination was carried out. Patients having cleft lip ⁄ palate associated with any history of developmental disabilities, including learning disabilities and attention deficit, hearing impairment, and speech deficit or abnormalities, were excluded from the study.
Inclusion criteria:
Exclusion criteria:
Blood samples (2 ml) were obtained from all subjects and stored with anticoagulant (EDTA), in an ice box. It was sent to the laboratory (Genohelix Biolab, Bangalore) within 24 hrs, for the SNP study.
Genomic DNA was extracted from the blood of the subjects by using ethanol–chloroform treatment. Once the isolated genomic DNA precipitate is obtained, it was used as a template for the polymerase chain reaction (PCR) test. The MSX1 (799 G>T) gene variant and MTHFR (677C>T) was determined, and the partial sequence of both the genes were amplified by PCR. The primers used in this study were procured from New England laboratories.
The MSX1 gene analysis was done using primer 2F (GGCTGATCATGCTCCAATGC) and a mutagenesis primer G267CR (CAGGAAACAGCTATGACCCTGGAAGGG GCCAGAGGCTC). The PCR product (448 bp) thus obtained was amplified. The amplified PCR products were digested with the specific restriction enzyme DdeI. PCR was carried out in a 20-µl volume containing 50 ng genomic DNA, 1X PCR buffer, 1.9–2.0 mM MgCl2, 0.2 mM dNTPs, 0.2 µM of each primer, and 0.5 U Taq polymerase, using the following parameters: 40 s at 94 ºC, 40 s at the annealing temperature and 40 s at 72 ºC for 35 cycles. The position of variants corresponds to the coding sequence for the nucleotide position within the Genbank entry AF426432.5
The MTHFR gene analysis was done using forward primer 5′-cca ccc cga agc agg gag ctt tg, and reverse primer 5′-tgg gaa aga tcc cgg gga cga tg. The PCR product (176 bp) thus obtained was amplified. The amplified PCR products were digested with specific restriction enzyme HinfI. The method adopted for PCR was restriction fragment length polymorphism (RFLP). The normal MTHFR nucleotide sequence was obtained with accession number U09806 in the GenBank database (National Institutes of Health, Bethesda, Maryland) at the website of the National Center for Biotechnology Information (http:// www.ncbi.nlm.nih.gov/ Genbank/index.html). Thermal cycling consisted of an initial denaturing step at 94ºC for 3 minutes, followed by 35 cycles at 94ºC for 30 seconds, annealing at 60ºC for 60 seconds, and primer extension at 72ºC for 60 seconds. A total of 10 µl of PCR product was added to 20 U of Hinf I, 2 µl 10 X RE buffer (New England Biolabs Inc., Beverly, Massachusetts), and double-distilled water was added to a final volume of 20 µl. After 3–4 hours of incubation at 37ºC, the reaction was stopped by heat inactivation at 80ºC for 20 minutes. The digests were then electrophoresed on a 3 percent Nusieve agarose gel (BioWhittaker Molecular Applications, Rockland, Maine) at 80 volts for approximately 1 hour. Hinf I-treated PCR products of known C677T genotypes and a sample of uncut PCR product served as controls and were run alongside samples being tested.2
The digested PCR products were separated into channels on a 1.5% agarose gel containing ethidium bromide in an electrophoretic chamber. An ultraviolet transilluminator was used to see the specific bands of base pairs of the digested PCR products.
RESULTS:
The search for the genetic cause of nonsyndromic cleft lip and palate has brought to focus the role of MSX1 and MTHFR gene in the development of Nonsyndromic clefting. As the linkage of a genetic disease is attributed to a defective gene (s), the presence of mutation in these genes would conclusively prove its role in nonsyndromic clefting of the South Indian patients.
Genomic DNA were isolated from 20 blood samples obtained from individuals with cleft lip and palate. All of the blood samples were successfully genotyped for both C677T polymorphism in MTHFR gene and G799T polymorphism in MSX1 gene.
The initial PCR product of the MTHFR gene was obtained for the twenty subjects. The size of this PCR product was 176 bp. Samples were then subjected to digestion with the specific restriction enzyme HinfI. Variant bands were seen after digestion in 13 subjects (Fig. 4).
The initial PCR product of the MSX1 gene was obtained for the twenty subjects. The size of this PCR product was 448 bp . Samples were then subjected to digestion with the specific restriction enzyme Dde1. No variant bands were seen after digestion. So the presence of variant was not shown by the twenty subjects.
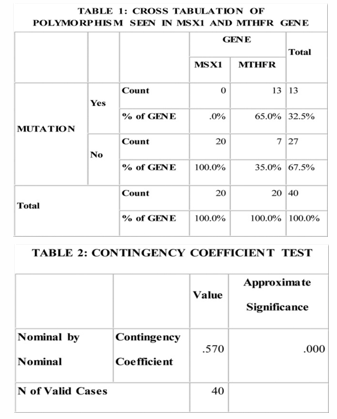
It was observed from Table 1 that polymorphism (gene mutation) was seen in MTHFR gene (65%) and not seen in MSX1 gene, for the present sample.
It was observed from Table 2 that compared to MSX1 Gene, the polymorphism in MTHFR gene was significant.
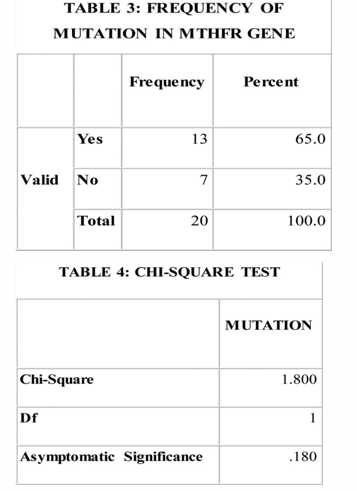
It was observed from Table 3 & 4 that, taking into consideration the polymorphism seen in MTHFR gene alone, the frequency of mutation was not significant.
DISCUSSION:
Both Cleft Lip and Palate (CLP) and Cleft Palate only (CPO) are caused by primary defects in the fusion of craniofacial processes that form the primary and secondary palate, respectively, in the first trimester of intrauterine development. Primary palate fusion takes place at about the fifth week of embryonic life by a highly regulated process of mesenchymal proliferation and epithelial breakdown or transformation in three facial prominences—the medial nasal, lateral nasal, and maxillary prominences—whereas elevation and fusion of the secondary palate occurs at about eight weeks.
Epidemiological and family studies indicate that CLP and CPO are separate etiological entities and for both multifactorial inheritance has been proposed. The precise roles played by genes and environment has not been elucidated; in particular, the nature and number of genes involved is not known. Whereas the role of heredity in the etiology of facial clefts, is well established, the contribution and exact role of exogenous factors, which also contribute to the etiology, remain uncertain.
Many variables have been examined in previous epidemiological studies, such as medication and drugs during pregnancy, illness, dietary deficiency, and environmental toxins; the effects of other variables such as sex, seasonal variation for conception, and birth weight have also been examined. Among these variables, maternal nutrition is thought to be important but has proved rather difficult to measure accurately and consistently, particularly around the time of conception and in the early stages of pregnancy. Nutritional supplements have proved to be instrumental in reducing the incidence of Neural Tube Defects, and recent evidence suggests that multivitamin or folic acid supplementation may reduce the risk of Orofacial Clefts.10 Both neural tube defects and oral clefts are thought to have a multifactorial inheritance, and occur more frequently together than can be expected by chance alone.8
In this study, we have evaluated 20 Nonsyndromic Cleft Lip/Palate patients for MTHFR and MSX1 gene poymorphism. 2 ml blood sample was taken using a 5 ml syringe and stored with anticoagulant (EDTA), in an ice box. It was sent to the Laboratory within 24 hrs, for the SNP study. Genomic DNA was extracted from the blood of the subjects by using ethanol–chloroform treatment. Once the isolated genomic DNA precipitate is obtained, it was used as a template for the polymerase chain reaction (PCR) test. The MSX1 (799 G>T) gene variant and MTHFR 677C>T was determined, and the partial sequence of the both gene were amplified by PCR. Restriction Fragment Length Polymorphism (RFLP) was done to check for variation in the MTHFR and MSX1 genes and the results were computed after statistical analysis.
Interpretation of the result
Graph 1
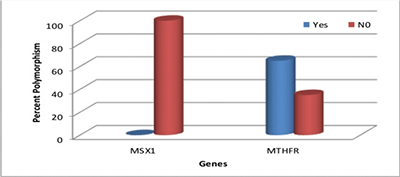
Graph 1 illustrates the percentage of MSX1 and MTHFR polymorphism seen in the sample. It shows that polymorphism seen in MSX1 gene was null and MTHFR gene was 65%. MTHFR gene polymorphism was statistically significant compared to MSX1 gene polymorphism.
Graph 2
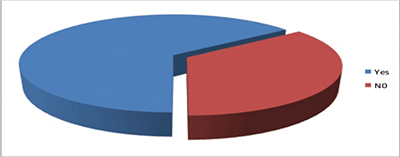
Graph 2 shows the proportion of MTHFR gene polymorphism seen in the given sample.65% of the subjects had gene polymorphism while 35% did not show any polymorphism. This result was not statistically significant enough to conclude on the role of MTHFR gene in clefting.
Graph 1: Percentage of MSX1 and MTHFR polymorphism in 20 subjects
Graph 2: Proportion of MTHFR polymorphism in given sample
It was inferred from the study that
Clinical implications
In every individual, genes form the basic framework for the development. Any minor changes or mutations in the structure of the gene can cause developmental anomalies or aberrations. Knowledge about the various genes can go a long way in preventing several abnormalities. The vast advances in technology and research in genetics now enable us to detect various genetic mutations in-uterine life. This can help detect and modify any aberrations present in the genes, thereby preventing developmental disturbances. Gene therapy in the future can and will play an important role in doing so.
Orofacial Cleft is a multifactorial disorder caused by a combination of genes and environmental interactions. These factors may contribute differently to Cleft Lip and/or Cleft Palate (CL/P) in different populations. Studies have shown that control of environmental factors helps reduce the risk of CL/P. Recent research might enable us to modify the aberrant genetic makeup of an individual, helping us prevent the occurance of CL/P. This requires the indepth knowledge of various genes responsible for CL/P. Through this study, it is seen that MTHFR gene, though not significant enough, does play a more prominent role than MSX1 gene in the development of Orofacial Clefts.
Somaiah et al.,Contribution of mthfr and msx1 to the risk of developing nonsyndromic cleft lip/palate- a snp study.2018:4(2);25-30
Subscribe now for latest articles, news.